Authors
Woojin Jung1, Koung Mi Kang2
1AIRS Medical Inc, Korea, Republic of
2Seoul National University Hospital, Korea, Republic of
[email protected]
KCR (2023)
PURPOSE
Brain volumetry is crucial for the diagnosis of neurodegenerative diseases. Although several MR-based volumetry products have been developed, standardization of image acquisition and validation of products for clinical use remain challenging issues. In particular, the accuracy of brain volumetry is significantly compromised when MR images are acquired with low quality. This study aimed to evaluate the effects of deep learning (DL)-based MR reconstruction in improving the accuracy of brain volumetry in low-quality MR images.
MATERIALS AND METHOD
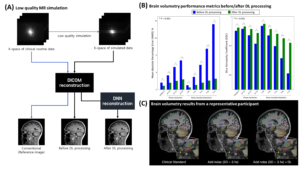
This retrospective study was approved by IRB. We collected 3D MPRAGE k-space data from 42 scans with the following scan parameters: TR/TE/TI = 1740/2.8/900 ms, voxel size = 1 x 1 x 1 mm3, GRAPPA = 2, and phase resolution = 0.7 ~ 0.8. From the k-space, we simulated 9 low-quality scans: one for GRAPPA = 4, three for varying phase resolutions from 0.4 to 0.6, and five for different levels of noise standard deviation (SD) from 1.4σ to 4.7σ, where σ is the noise SD of original k-space. Each simulated k-space was reconstructed into DICOM images. Brain volumetry was performed using Fastsurfer (ver 2.0.0), resulting in 96 sub-region segmentation maps. In each sub-region, two error metrics, the mean absolute percentage error (MAPE) of the estimated volume and dice similarity coefficient (DSC), were calculated with the original scans as reference. To evaluate the effects of DL-based MR reconstruction, we applied the software (SwiftMR™, AIRS Medical) that functions to denoise and achieve super-resolution of MR images to all DICOMs. Then, the same brain volumetric analysis was repeated. The results before and after DL processing were compared by a paired t-test.
RESULTS
In all low-quality simulations, the MAPE and DSC values were significantly improved in DL-processed images. For example, error metrics before and after DL were estimated at (MAPE, DSC) = (1.6%, 0.95) vs. (0.9%, 0.96) in GRAPPA of 4; (MAPE, DSC) = (6.8%, 0.91) vs. (2.4%, 0.94) in phase resolution of 0.4; and (MAPE, DSC) = (15.2%, 0.81) vs. (2.5%, 0.91) in noise SD of 4.7σ, all P < 0.001. Brain volumetric results of example slides supported the improvement by DL processing.
CONCLUSION
DL-based MR image reconstruction significantly improved the accuracy of brain volumetry in low-quality MR images. This approach has the potential to address issues related to image quality standardization and enable the clinical use of brain volumetry, even allowing for the comparison of images from different MRI scanners.

